 Protein production
Protein production
A guide to recombinant protein expression: hosts, vectors & tags
Recombinant protein expression is considered one of the most important breakthroughs ever achieved in the life sciences. Decades of research have led to the continuous refinement of the protein expression process and diversification of expression systems and hosts, vectors, and promoters. In practical terms, the vast choice may often hamper the decision-making process. To guide you through this process and help you make informed decisions, we have summarized all key properties, advantages, and limitations of each approach to protein expression on this page.
- Mammalian protein expression – CHO and HEK293
- Get more out of your transient system
- 5 smart ways XtenCHOTM improves your yields without sacrificing your time
- Insect protein expression – Sf21, Sf9, and BTI-TN-5B1-4 (High Five)
- Yeast protein expression – Saccharomyces cerevisiae and Pichia pastoris
- Bacterial protein expression – Escherichia coli and Bacillus subtilis
- How to choose an expression system
- Basic design of a protein expression vector
- Common promoters used for protein expression
- Common tags used for protein expression
- Common selectable markers used for protein expression
- Transformation, transduction, and transfection reagents and methods
Mammalian protein expression – CHO and HEK293
Mammalian systems are considered one of the best for protein expression due to their enhanced protein folding, ultra-high yields, and ability to perform human-like glycosylation, which is vital for therapeutic proteins such as antibodies. A wealth of knowledge is currently available regarding the genetic background, promoters, transfection reagents, and protein purification methods used for mammalian systems. Additionally, a wide range of cell lines, growth media, and expression vectors are also commercially available, making it easy to optimize expression in these systems by allowing researchers to screen several constructs in parallel for optimal results.
Because mammalian cells are unable to replicate plasmids independently, two different methodologies were developed for these systems – stable and transient. Stable expression requires the total integration of the gene of interest into the host’s genome via recombination or transposon-mediated gene transfer. Due to the time-consuming nature of this methodology, stable cell line generation is typically reserved for large-scale production. In contrast, transient expression harnesses the superior features of mammalian systems while considerably reducing lead times. These properties make them highly desirable for research and analytical applications, and drug and protein variant screening purposes.
The typical steps required for protein expression in transient mammalian systems include:
- Preparation of coli competent cells (via the use of chemical or electrical signals)
- Codon optimization and synthesis of the gene of interest
- Cloning of the gene of interest into the expression vector
- Transformation of coli cells
- Selection and expansion of positive clones
- Isolation of plasmid DNA
- Transient transfection of mammalian cells
- Expansion of the transient batch for protein production and purification
Typical mammalian cell lines used for recombinant protein production include:
- Chinese hamster ovary (CHO)-derived cell lines used for protein expression include CHO-K1, CHO-S, CHO-DXB11, and CHO-DG44
- Human embryonic kidney (HEK)-derived cell lines used for protein expression include HEK293T and HEK293E
More than 70% of all biopharmaceuticals are currently produced in CHO cell lines. These genes are easy to handle, amenable to high-density cultures, and safer than HEK which, due to its human origin, may harbor and disseminate human pathogens.
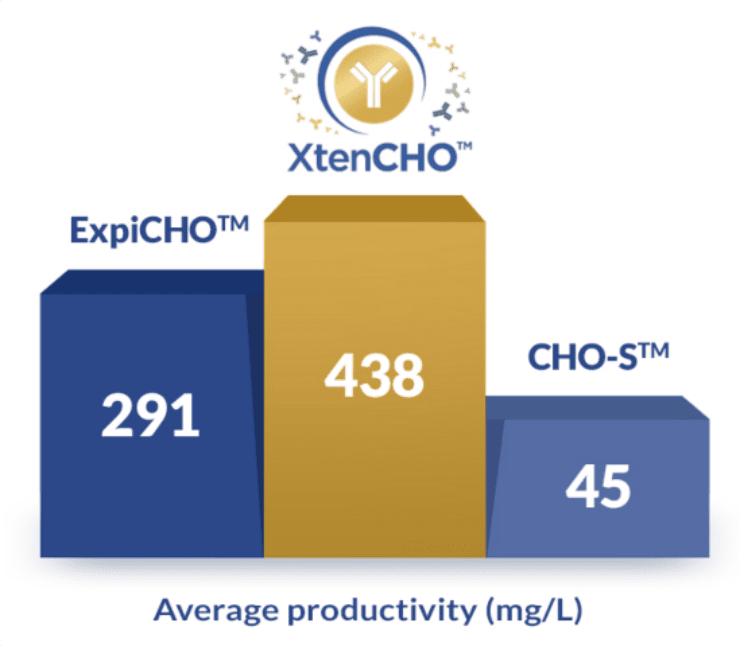
Get more out of your transient system
5 smart ways XtenCHOTM improves your yields without sacrificing your time
- 10x higher yields without supplemental feeds
- Smarter protocols with a single one-step transfection
- 3x longer transient expression periods
- 1,500+ proteins successfully produced
- Flexible purchase options adapted to your needs (buy the starter kit, cell, and medium separately only when you need them)
Insect protein expression – Sf21, Sf9, and BTI-TN-5B1-4 (High Five)
Protein expression in insect cells is comparable in terms of quality and yield to mammalian systems. Insect cells are capable of performing highly efficient protein folding, post-translational modifications, and protein trafficking, all in shorter lead times than mammalian systems. However, glycosylation pathways in insects differ significantly from higher eukaryotes, making them less optimal for the production of full-length therapeutic antibodies.
This expression system is commonly used when specific proteins fail to be expressed in bacterial and yeast systems. Currently, the most popular form of insect cell expression is mediated by baculoviruses, arthropod-specific viruses. Interestingly, these viruses can be adapted to propagation in Escherichia coli. These hybrid vectors are named bacmids and represent a highly cost-effective solution for recombinant protein expression.
The typical steps required for protein expression in insect/baculovirus systems include:
- Preparation of coli competent cells (via the use of chemical or electrical signals)
- Codon optimization and synthesis of the gene of interest
- Cloning of the gene of interest into the expression vector (bacmid)
- Transformation of coli cells
- Selection and expansion of positive clones
- Isolation of plasmid DNA
- Transfection of insect cells with the expression vector (bacmid) and a second plasmid containing vital genes for the formation and multiplication of viral particles
- Purification of the recombinant viral stock
- Amplification of the virus
- Infection of insect cells with the recombinant virus
- Protein production, scale-up, and purification
Typical insect cell lines used for recombinant protein production include:
- The Sf21 cell line (IPLB-Sf21-AE) was established from ovarian cells of the Fall Armyworm (Spodoptera frugiperda) a species of the order Lepidoptera (moths and butterflies)
- The Sf9 cell line was developed from a clonal isolate of the Spodoptera frugiperda Sf21 cell line
- High Five cells (BTI-TN-5B1-4) were first established from ovarian cells of the cabbage looper Trichoplusia ni from the order Lepidoptera – it is one of the most popular cell lines currently used for recombinant protein expression using baculovirus
Cell lines S2 and S2R+ from Drosophila melanogaster of the order Diptera (flies).

Discover our insect/baculovirus protein expression services
Yeast protein expression – Saccharomyces cerevisiae and Pichia pastoris
Yeast cells are the simplest eukaryotic protein production systems. They are uniquely poised between the enhanced protein folding and modification capacities of mammalian systems and the simplicity and flexibility of bacterial systems. Although they are unable to perform human-like glycosylation required for some pharmaceutical proteins such as antibodies, yeast systems are prized for their fast growth, ease of genetic manipulation, scalable fermentation, safe pathogen-free production, and high production yields.
An attractive advantage of yeast vectors is their ability to replicate autonomously in host cells (episomal vectors). This feature considerably shortens the development timeline because it forgoes the need to stably integrate the foreign genes into the host’s genome. Episomal vectors provide high copy numbers and high protein yields; however, they are easily lost in the absence of selection. Transformation of yeast systems is also simplified because yeast vectors are easily produced in Escherichia coli (shuttle vectors) before being propagated in the final host.
The typical steps required for protein expression in yeast include:
- Preparation of coli competent cells (via the use of chemical or electrical signals)
- Codon optimization and synthesis of the gene of interest
- Cloning of the gene of interest into the expression vector
- Transformation of coli cells
- Selection and expansion of positive clones
- Isolation of plasmid DNA
- Transfection of yeast cells using the coli-amplified shuttle vector
- Selection and screening of positive clones (stable integration or episomal vector)
- Expansion of positive yeast clones
- Protein production, scale-up, and purification
Typical yeast strains used for recombinant protein production include:
- Non-methylotrophic yeast Saccharomyces cerevisiae strains BJ5464, AH22, and INVSc1 are the most widely used for recombinant protein production
- Methylotrophic yeast Pichia pastoris strains GS115, KM71, and X‐33 are the most widely used for these applications
P. pastoris has been gaining ground over more conventional S. cerevisiae systems for protein expression due to its ability to achieve higher cell densities and superior protein yields.

Discover our yeast protein expression services
Bacterial protein expression – Escherichia coli and Bacillus subtilis
Bacteria are one of the most widely used expression systems due to their inexpensive requirements, fast growth, ease of genetic manipulation, straightforward scale-up process, and high protein yields. From a practical point of view, bacteria serve as excellent starting points for many functional studies and research applications. Plus, many biopharmaceuticals are currently produced in bacteria such as Escherichia coli.
However, this expression system remains limited in terms of post-translational modifications and protein folding, making it impractical for the production of complex proteins such as IgG-like antibodies (monoclonal or bispecific). Bacterial systems also tend to produce proteins intracellularly requiring harsh extraction and purification methods. Additionally, some proteins may be produced as insoluble inclusion bodies, requiring the use of denaturants and subsequent protein-refolding processes, substantially reducing purity and yields.
The typical steps required for protein expression in bacteria are the following:
- Preparation of competent cells (via the use of chemical or electrical signals)
- Codon optimization and synthesis of the gene of interest
- Cloning of the gene of interest into the expression vector
- Transformation of competent cells (i.e., heat shock or electroporation)
- Selection of positive clones by growing in selective medium
- Expansion of positive clones
- Protein production, scale-up, and purification
Typical bacterial strains used for recombinant protein expression:
- Escherichia coli strains BL21(DE3) and derivates, strains of the K-12 lineage (e.g., HMS147(DE3)), and strain AD494 are the most widely used for protein expression
- Bacillus subtilis strains WB600 and WB800
Substantially more information, plasmids, and strains are available for protein expression in E. coli. The use of B. subtilis is limited due to the lack of standardized expression vectors and systems based on this species. Although this gram-positive bacterium presents some advantages over E. coli such as the ability to secrete proteins to the growth medium, much work remains to be done before this system reaches commercial maturity.

Discover our protein expression services in E. coli and B. subtilis
How to choose an expression system
Bacteria, yeast, insect cells, and mammalian cells are all widely used systems for recombinant protein production. Each system has its unique advantages and limitations:
| ADVANTAGES | LIMITATIONS | |
|---|---|---|
| Mammalian systems | · Enhanced protein folding
· Human-like glycosylation · Efficient protein secretion – cost-effective purification · High batch-to-batch consistency · High purity · High production yields meeting important quality standards · Adapted to most types of proteins · Fast production possible (transient) |
· Longer lead times (stable)
· Time-consuming and laborious (stable) · Cell lines are harder to engineer due to the complexity of their genomes · Need for incubators with CO2 gassing to maintain suitable pH values · Risk of viral infections or contamination with mycoplasma |
| Insect systems | · Good production yields
· Adequate protein folding · Ability to carry out complex post-translational modifications – comparable to mammalian systems · Production is easily scaled up |
· Time-consuming and laborious process due to the need to produce baculovirus (bacmid) for transduction
· Risk of cell lysis upon infection – may reduce protein purity and quality |
| Yeast systems | · Efficient protein secretion – easier and cost-effective purification
· Ease of genetic manipulation · Short turnaround times · Ability to carry out some post-translational modifications · No stable integration required for large-scale production · Production is easily scaled up |
· Moderate risk of plasmid loss even in selective conditions
· Inability to perform human-like glycosylation without extensive engineering · Yeast cells may be difficult to lyse resulting in protein denaturation during extraction and the need for in vitro refolding |
| Bacterial systems | · Extremely short turnaround times
· Advantageous for antibody fragment (scFv, Fab, VHH) and small protein production · Ease of genetic manipulation · No stable integration required for large-scale production · Production is easily scaled up · Extremely cost-effective · Inexpensive medium and growth conditions required |
· No post-translational modifications
· Intracellular production – harsher purification methods required · Risks of forming inclusion bodies leading to low yields · Contamination with endotoxins (E. coli) requiring additional purification steps when proteins are meant for biopharmaceutical or cell culture use |
Despite the different advantages and limitations of these systems, it is important to bear in mind that choosing an expression depends, first and foremost of:
- The origin of the target protein – bacterial proteins are better expressed in bacterial systems, while mammalian proteins are better expressed in mammalian systems. For this reason, mammalian cell lines like CHO (Chinese Hamster Ovary) or HEK (Human Embryonic Kidney) are the systems of choice for human protein production
- Required post-translational modifications – proteins devoid of complex modifications (e.g., glycosylation, alkylation, phosphorylation, or specific proteolytic processing) are more easily produced in simpler systems with short turnaround times such as bacterial and yeast. In contrast, complex proteins with significant modifications should be produced either in insect or mammalian cells
- Solubility of the recombinant protein – some proteins are not properly folded in bacterial systems, as a result, they tend to form insoluble aggregates (inclusion bodies) that are difficult to extract. In these circumstances, higher eukaryotic systems should be preferred for the recombinant production of the target protein
The intended application of the recombinant protein – even complex proteins such as IgG-like antibodies may be produced in simpler with high turnover systems. In these proteins, glycans are restricted to the Fc fragment (crystallizable fragment) and modulate the effector functions of these biomolecules. However, even devoid of glycans, antibodies can bind to target antigens. For this reason, antibodies for research (functional studies, medical, or basic research) and in vitro diagnostics can be produced in bacteria or yeast, because effector functions are non-essential for these applications. In contrast, antibodies intended for therapeutic use are mostly produced in mammalian systems able to perform human-like glycosylation essential to modulate the immune response.
XtenCHO transient expression
Get the most out of your transient expression experiments
with XtenCHO. Ultra-high yields, minimized hands-on time,
and longer transient expression periods.
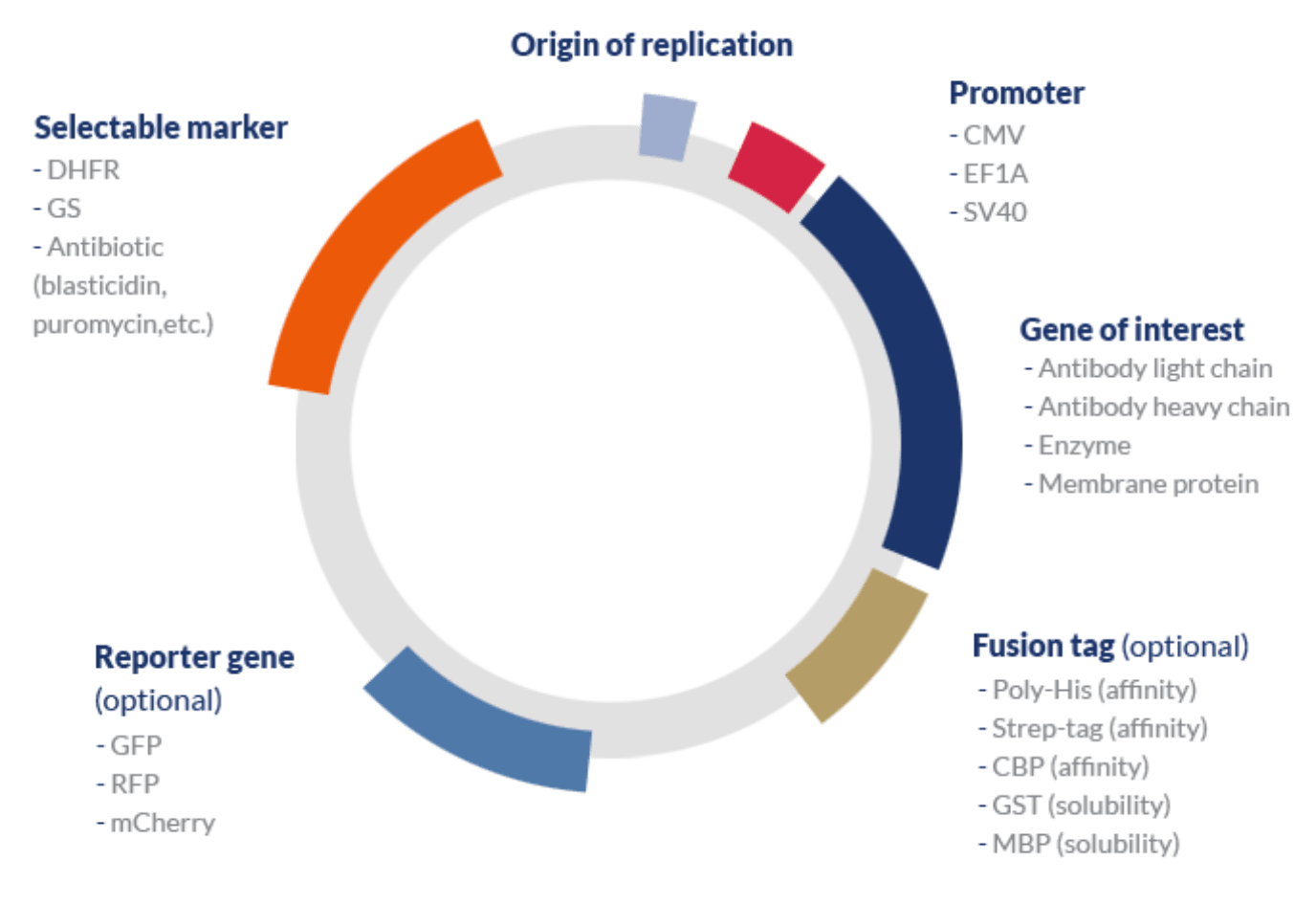
Basic design of a protein expression vector
A typical expression vector used in mammalian systems may include the following elements:
- One or several genes of interest (GOI) associated with a strong inducible promoter and a transcription terminator.
- Bacterial origin of replication (ori) – although mammalian systems are typically unable to recognize bacterial origins or naturally replicate plasmids, this element is used during the process of cloning the GOI into the vector and amplifying the construct. A step typically carried out in hosts that are easy to handle such as coli.
- A selectable marker – mammalian cell selection can be carried out via antibiotic or metabolic selection.
- Reporter gene – because transient systems rarely undergo a process of single clone selection, the efficiency of the transfection process can be assessed by measuring the expression of a specific reporter gene. This gene typically encodes fluorescent proteins such as GFP (green fluorescent protein) that can be easily be measured via non-destructive methods.
- Tags – affinity or solubility tags are typically included in mammalian expression vectors to improve the stability of the protein of interest and enhance the efficiency of the purification process, respectively.
- Optional elements – signal peptides can be used to improve translocate the protein of interest to the cell membrane and enhance protein secretion; protease recognition sites can be used to allow the cleavage of the peptide link between tags and the protein of interest; among others.
Common promoters used for protein expression
Promoters modulate the transcription process by controlling the binding of the RNA polymerase and associated factors to the DNA chain. This element dictates the timing and amount of protein that will be produced by a specific recombinant system. These elements are generally classified as constitutive, inducible, or repressible.
The first type of promoters is always active independently of the circumstances. In contrast, inducible promoters are only activated by specific signals, acting like an ON switch. Repressible promoters have found limited applicability in recombinant protein production. However, they are substantially used in research. In their basal state, these promoters are always active, but they can be repressed by specific signals. Thus, these promoters act as OFF switches.
In the table below, you can find a list of the most commonly used promoters in both eukaryotic and prokaryotic recombinant expression systems:
| Promoter | Origin | Promoter type | |
|---|---|---|---|
| EURAKYOTIC | CMV | Human cytomegalovirus | Constitutive |
| EF1A | Human elongation factor alpha | Constitutive | |
| TEF1 | Yeast transcription elongation factor (analogous to EF1a) | Constitutive | |
| SV40 | Simian vacuolation virus 40 | Constitutive | |
| UBC | Human ubiquitin C gene | Constitutive | |
| PGK | Human or mouse phosphoglycerate kinase gene | Constitutive | |
| CAG | Synthetic promoter constructed from CMV, chicken beta-actin gene, and rabbit beta-globin gene | Constitutive | |
| Ac5 | Drosophila Actin 5c gene | Constitutive | |
| OpIE 1 & 2 | Orgyia pseudotsugata multicapsid nuclear polyhedrovirus | Constitutive | |
| PH | Baculovirus promoter (polyhedrin) | Constitutive | |
| P10 | Baculovirus promoter | Constitutive | |
| GAP | Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene | Constitutive | |
| TET | E. coli Tn10-specified tetracycline-resistance operon | Inducible (tetracycline) | |
| GAL4/UAS | Yeast transcriptional activator GAL4 and upstream activation sequence UAS | Inducible (heat shock) | |
| cTA | p-cmt and p-cym operon in Pseudomonas putida | Inducible (cumate) | |
| MT | Drosophila metallothionein promoter | Inducible (copper) | |
| AOX1 | Pichia pastoris alcohol oxidase gene AOX1 | Inducible (methanol) | |
| GAL1 | Saccharomyces cerevisiae galactokinase gene GAL1 | Inducible (galactose) | |
| ADH1 | Yeast promoter for alcohol dehydrogenase I | Repressible (ethanol) | |
| PROKARYOTIC | T7 | T7 bacteriophage promoter | Constitutive* |
| T7lac | T7 bacteriophage promoter + lac operators | Constitutive* | |
| Sp6 | Sp6 bacteriophage promoter | Constitutive* | |
| T3 | T3 bacteriophage promoter | Constitutive* | |
| lac | Promoter from the lactose operon | Inducible (IPTG) | |
| Ptac | Hybrid synthetic promoter of lac and trp | Inducible (IPTG) | |
| pL | Promoter from bacteriophage lambda | Inducible (temperature) | |
| araBAD | Promoter of the arabinose metabolic operon | Inducible (arabinose)
Repressible (glucose) |
|
| trp | Promoter of E. coli tryptophan operon | Repressible (tryptophan) |
*Require T7, Sp6, or T3 RNA polymerase, respectively. Highlighted in grey are the most commonly used promoters.
Inducible promoters are the standard among prokaryotic expression systems. In contrast, constitutive promoters are dominant among mammalian and other eukaryotic systems. They can deliver high-level expression without the need to use additional signals (i.e., chemicals, heat, etc.), which are often toxic, expensive, or known to affect the integrity of the mammalian cell. This uncontrolled expression becomes disadvantageous only when expressing toxic proteins for long periods of time. In these specific circumstances, inducible or repressible promoters can be used to minimize the risks of early cell death and maintain cell viability for as long as possible.
Common tags used for protein expression
Fusion tags can be short peptide sequences or large proteins. They are recurrently used to improve purification (affinity or epitope tags), solubility (solubility tags), or simply as a means to monitor or visualize protein expression (fluorescent tags). Tags can be removed by chemical or enzymatic methods. However, this is only done if the activity or structure of the recombinant protein are severely affected. In fact, many tags were found to improve stability during storage and extend the shelf-life of recombinant proteins.
Large tags are more commonly used to improve solubility, while shorter tags are more commonly used to mediate protein purification processes. In comparison, affinity tags are also larger than epitope tags. The first ones are fused to the protein of interest, allowing the purification via affinity chromatography. While epitope tags are typically engineered into the protein of interest, allowing antibody-mediated purification. Both affinity and antibody-mediated purification processes can be carried out by immobilizing antibodies or other compounds on resins (affinity purification) or on magnetic beads (magnetic-based purification). The former is suitable for large volumes, while the latter is more suitable for small-scale purification.
| Tag name | Type | Sequence or Size | Matrix |
|---|---|---|---|
| Hemagglutinin (HA) | Epitope | YPYDVPDYA (9) | Anti-HA antibody |
| c-myc | Epitope | EQKLISEEDL (10) | Anti-Myc antibody |
| V5 | Epitope | GKPIPNPLLGLDST (14) | Anti-V5 antibody |
| FLAGTM | Epitope | DYKDDDDK (8) | Anti-FLAG antibody |
| Poly-His | Affinity | nH (n=2-12, usually 6) | Metal ions |
| Poly-Arg | Affinity | nR (n=5-6, usually 5) | Cation exchange resin |
| Strep-tag | Affinity | WRHPQFGG (8) | Streptavidin |
| Strep-tag II | Affinity | WSHPQFEK (8) | StrepTactin |
| Twin-Strep-tag | Affinity | 3 kDa | StrepTactin |
| Biotin acceptor protein (BAP) | Affinity | GLNDIFEAQKIEWHE (15) | Avidin |
| Calmodulin-binding protein (CBP) | Affinity | 3 kDa | Calmodulin |
| Chitin-binding tag | Affinity | 6 kDa | Chitin |
| Maltose-binding (MBP) tag | Solubility | 43 kDa | Transversely-linked amylose |
| Glutathione-S-transferase (GST) tag | Solubility | 27 kDa | Glutathione |
| SUMO tag (small ubiquitin-like modifier) | Solubility | 12 kDa | Accommodates any type of affinity tags such as 6xHis |
| NusA tag | Solubility | 55 kDa | Accommodates any type of affinity tags such as 6xHis |
| TrxA | Solubility | 24 kDa | Accommodates any type of affinity tags such as 6xHis |
| Fh8 | Solubility | 8 kDa | Accommodates any type of affinity tags such as 6xHis |
| Halo | Solubility | 33 kDa | Chloroalkanes |
| Green Fluorescent Protein (GFP) | Fluorescence | 27 kDa | Anti-GFP antibody or fusion with an affinity tag |
| Red Fluorescent Protein (RFP) | Fluorescence | 26 kDa | Anti-RFP antibody or fusion with an affinity tag |
| mCherry | Fluorescence | 28 kDa | Anti-mCherry antibody |
Tag names shown in bold represent the most commonly used fusion tags.
Of all tags mentioned, it is worth mentioning that fluorescent tags are extremely useful when working with transient mammalian systems. Because transient systems tend to lose their expression vectors with each cell division cycle, it is not recommended to select and isolate positive clones before protein production. In these systems, high transfection efficiencies directly correlate with high protein titers. For this reason, researchers use fluorescent tags to measure and optimize transfection efficiency.
Affinity and epitope tags are recurrently used on all expression systems. Alternatively, recombinant antibodies can be purified using the following methods:
- Immunoglobulin-specific purification: a wide range of microbial proteins is known to bind strongly to antibody’s Fc or Fab fragments. A few examples are protein A (Staphylococcus aureus), protein G (Streptococcus), fusion protein A/G, and protein L (Peptostroptococcus magnus). These proteins can be immobilized into resins and easily used to capture antibodies from the culture medium.
- Antigen-specific purification: alternatively, antibodies can be captured by immobilizing their specific antigen in a resin.
Interestingly, these purification methods are typically reserved for small and medium-scale antibody production because they can become expensive at the large-scale. To reduce purification costs at this scale, antibodies can also be associated with a number of different affinity or epitope tags.
Common selectable markers used for protein expression
Positive selection is typically carried out via metabolic or antibiotic selectable markers. Metabolic selection is generally used in mammalian systems while antibiotic-mediated selection is the standard method for bacterial cell selection.
| Type | Name | Gene | Selection |
|---|---|---|---|
| Metabolic selection (eukaryotic) | DHFR system | Dihydrofolate reductase gene (dhfr) | Used with DHFR- cell lines or cells pre-treated with methotrexate (MTX), a powerful DHFR inhibitor. Selection is made in media devoid of glycine, hypoxanthine, and thymidine. |
| GS system | Glutamine synthetase gene (glul) | Used with GS- cell lines or cells pre-treated with methionine sulfoximine (MSX), a GS inhibitor. Selection is made in the presence of MSX and glutamine – only degraded by GS+ cells. | |
| Antibiotic (eukaryotic) | Blasticidin | Blasticidin-S deaminase gene (bsd) | Protein synthesis inhibition by binding to the ribosome |
| Hygromycin B | Hygromycin B phosphotransferase gene (hygB) | Protein synthesis inhibition by binding to the ribosome | |
| G418/Geneticin | Neomycin-resistance gene (neo) | Protein synthesis inhibition by interfering with initiation and elongation | |
| Puromycin | Puromycin N-acetyltransferase (pac) | Protein synthesis inhibition by blocking the entrance of aminoacyl-tRNA | |
| ZeocinTM | ZeocinTM resistance gene (Sh ble) | Causes cell death by intercalating with DNA and cleaving it via reactive radicals | |
| Antibiotic (prokaryotic) | Ampicilin | Beta-lactamase (amp) | Inhibition of the cell wall synthesis |
| Chloramphenicol | Chloramphenicol acetyltransferase (cat) | Protein synthesis inhibition by binding to the bacterial 50S ribosomal subunit | |
| Kanamycin | Neomycin phosphotransferase (kan) | Protein synthesis inhibition by interfering with initiation and elongation | |
| Tetracycline | Tetracycline resistance protein (tetA) | Protein synthesis inhibition by binding the bacterial 30S ribosomal subunit |
Transformation, transduction, and transfection reagents and methods
Transformation, transduction, and transfection are used to describe the introduction of foreign DNA or RNA into eukaryotic or prokaryotic cells. Transformation and transduction occur naturally and are collectively named horizontal gene transfer (HGT) processes. In contrast, transfection is a man-made protocol used to insert genes into eukaryotic cells (mainly mammalian).
Transformation is the process through which bacteria assimilate genetic material from their surrounding environment. It occurs spontaneously and frequently in nature, often promoted by the presence of specific substances. In the lab, researchers harness the process of natural transformation by:
- Preparing competent cells – to increase the efficiency of natural transformation processes, bacterial cells can be treated with specific chemicals or electric shocks to make their membranes more permeable to genetic material.
- Transforming competent cells – bacterial cells are typically transformed by mixing competent cells with naked plasmid DNA followed by heat shock or electroporation.
Transduction occurs when foreign genetic material is introduced into prokaryotic or eukaryotic cells via a virus or viral vector. Viruses interact with cells via specific membrane-bound receptors. As a result, they undergo important structural changes leading to the injection of their genomes (DNA or RNA) into the cytoplasm of their host.
Within the cell, the genetic machinery hijacks the cellular processes to produce multiple copies of the virus. Alternatively, the extrachromosomal DNA or RNA is integrate the host’s genome through a process of homologous or site-directed recombination. Transduction can be used in all different expression systems. However, it is more commonly applied to insect systems.
Mammalian cells are unable to uptake naked plasmid DNA, they can only be modified via transduction or transfection. Transduction is generally considered an expensive and time-consuming process, especially when it comes to mammalian systems. The reason stems from the need to produce the viral vectors in other mammalian cells. For this reason, protein expression in these systems is conventionally carried out via transfection using chemical or physical methods.
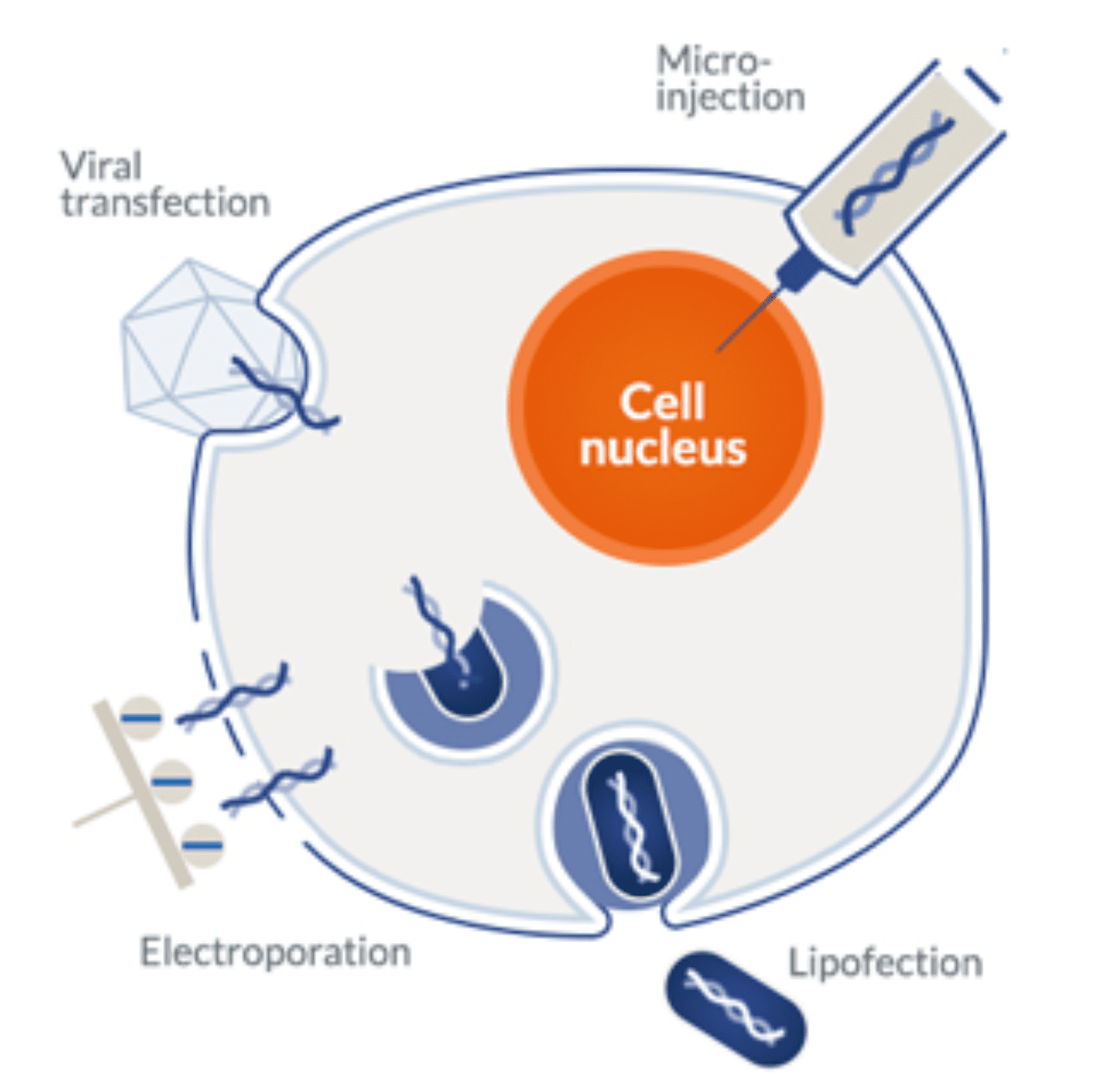
Chemical methods remain the most popular and widely used approach of the two due to the ease, low cost, and wide variety of commercial transfection reagents. The most common transfection methods used in mammalian systems include:
- Chemical methods
- DEAE-Dextran: diethylaminoethyl (DEAE)-dextran is a polycation derived from dextran. Due to its cationic nature, DEAE associates naturally with negatively charged nucleic acids. The resulting complex adheres easily to cell membranes, and the foreign DNA or RNA are thus free to enter the cytoplasm via endocytosis. This method was one of the first to be developed for mammalian transfection. It is relatively simple, with high reproducibility and cost. However, its limitations include cytotoxicity, low transfection efficiency, and the requirement to reduce the medium during the transfection process.
- Calcium phosphate: this method is based on the formation of calcium phosphate-DNA precipitates. Calcium mediates the binding of the complex to the cell membrane and the foreign DNA enters the cell by endocytosis. It represents one of the first methods used for mammalian transfection. Currently, its use remains limited due to the inherent toxicity of this reagent and poor transfection efficiency.
- Lipotransfection: liposomes are synthetic analogs of the phospholipid bilayer, for this reason, they fuse easily with cell membranes. Liposomes encapsulate free DNA and RNA molecules quite efficiently, thus forming a simple delivery system able to introduce these nucleic acids into the cytoplasm by endocytosis. These are considered first-generation chemical transfection methods and are only sparsely used since the development of newer, more effective reagents.
- Cationic lipid transfection: this method leverages the ease of fusion of liposomes with cationic polymers’ natural ability to form strong complexes with negatively charged nucleic acids (DNA or RNA). Like first-generation reagents, these are taken up by the cell via endocytosis and subsequently release their cargo into the cytoplasm. Exogenous DNA is further translocated to the cell nucleus to be transcribed, while RNA-based vectors stay in the cytoplasm for direct translation.
- Physical methods
- Electroporation: this method uses electrical pulses to create temporary pores in cell membranes. This process makes membranes permeable to negatively charged nucleic acids and allows them to migrate directly into the cytoplasm without the need for additional reagents. Electroporation typically results in higher transfection rates. However, it is more labor-intensive and harder to optimize than conventional chemical methods. One of the most important drawbacks of this method is the substantial cell death caused by high voltage pulses. For this reason, this method also requires a greater quantity of cells in comparison to chemical methods.
Microinjection: as the name indicates, microinjection refers to the process of injecting foreign DNA or RNA into the cytoplasm of a cell via a very thin needle. Despite the high degree of control and efficiency achieved via microinjection, it has a very low throughput. For this reason, it is mostly reserved for single-cell applications.
XtenCHO transient expression
Get the most out of your transient expression experiments
with XtenCHO. Ultra-high yields, minimized hands-on time,
and longer transient expression periods.