Blog
Blog
Which approach should you choose to get a species-specific veterinary monoclonal antibody? A decision guide
The right antibody development approach depends on two factors: your starting point and your target species. Use this guide's decision tree and provider checklists to map your specific path forward.
Developing monoclonal antibodies for veterinary applications presents unique challenges. Unlike human medicine’s single-species focus, veterinary biologics must work across diverse species, from companion animals to livestock, each with distinct immunological considerations.
The veterinary antibody market is growing, with successful products like Cytopoint® (Lokivetmab, anti-canine IL31, USDA-approved in 2016) demonstrating the potential. However, obtaining antibodies that are both effective and species-compatible remains a critical bottleneck.
Whether you’re adapting an existing antibody to a new species or starting discovery from scratch, choosing the right approach impacts your timeline, budget, and success rate. This guide breaks down your options based on your starting point, covering traditional methods and emerging AI-driven solutions to help you navigate the path to a species-specific veterinary monoclonal antibody.
Case n°1: You have an antibody not adapted to your target species
If you already have your lead antibody, whether it’s a commercial one or your own development project, you will have to perform speciation.
Speciation consists of adapting an antibody from species A to species B, in order to reduce as much as possible its immunogenicity in species B.
However, this speciation process should not significantly impact your antibody’s performance. This delicate balance of reducing immunogenicity while preserving characteristics such as affinity or specificity, is the main challenge of speciation.
Traditional methods like CDR-grafting have proven their efficiency for species switch. Nevertheless, this method often requires additional affinity maturation as it is more likely to disrupt the antibody’s performance.
AI-powered speciation
In the last few years, we have witnessed the emergence of AI in biology. This powerful computational tool can now be used for speciation. One advantage is that AI models can incorporate developability concerns to deliver species-specific antibodies with unchanged or even improved performance and manufacturability.
Tip: However, it is not systematically the case. Make sure to ask your provider if you plan on outsourcing this part.
Learn more about our AI-driven speciation service
AI speciation presents other advantages. It is designed to provide the most likely to succeed species-specific sequences. Therefore, fewer sequences need to be tested in wet-lab, which reduces cost and time.
Tip: If you plan on outsourcing, check that your provider includes wet-lab testing and guidance to interpret the AI’s results. Be careful to avoid black box AI services.
Checklist: Questions to ask a service provider before outsourcing
- Quality of the AI database: How many sequences are in the database? → A high-quality database should contain billions of sequences
- Experience: Does the provider have experience in species switch? → Make sure the provider does not only do humanization
- Guidance: Do they provide personalized guidance from bioinformatic experts? → To have relevant guidance, you must be advised by people specialized in AI informatics and biology.
- Developability: Does their AI model include affinity and developability improvements? → The provider should at least be prepared to conserve the current performance, but we advise doing the developability improvement step at the same time to save time and money.
Case n°2: You do not have an antibody
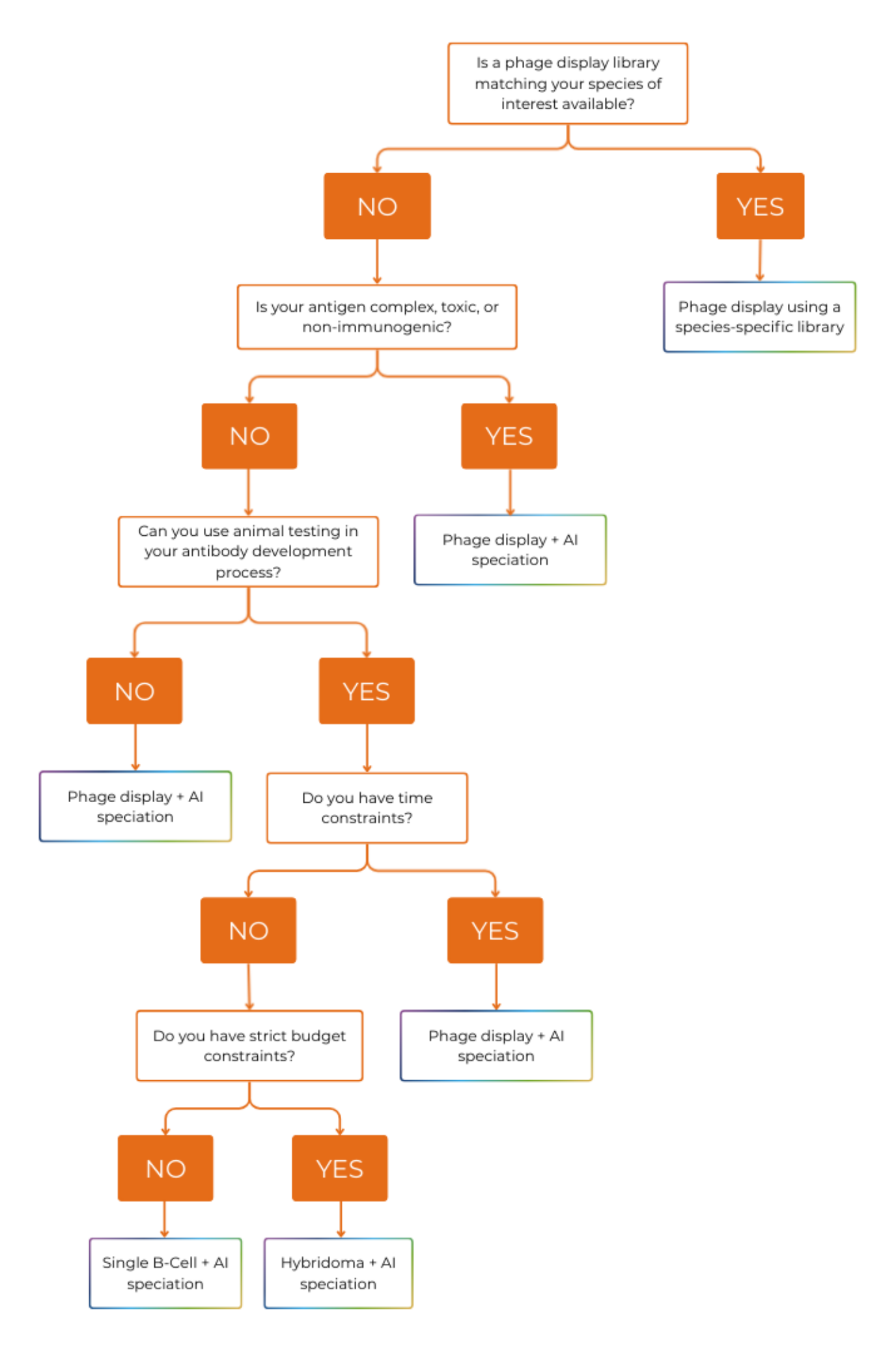
If you do not have an antibody to start with, the most direct method is phage display using a species-specific library. This discovery method is very convenient as it provides antibodies already corresponding to your target species. After discovery, some developability improvement or reformatting may be needed according to your project, but it is still the most convenient method.
Nevertheless, libraries corresponding to less common species may be hard or impossible to find. If you plan on outsourcing, some service providers only offer human libraries or small ones for pets (usually dog, cat and rabbit). Cattle libraries are much rarer.
See our pets & cattle libraries
Therefore, if you cannot find a phage display library adapted to your species of interest, you need to dissociate discovery and speciation. We recommend AI speciation only with a suitable partner (use our checklist for service provider choice). Otherwise you can rely on CDR grafting.
To help you choose the complementary discovery method, we made a decision tree to find the best option according to your project.
Explore our discovery services:
Bibliography
Apgar, J. R., Mader, M. M., Agostinelli, R., Benard, S., Bialek, P., Johnson, M., Gao, Y., Krebs, M., Owens, J., Parris, K., St Andre, M., Svenson, K., Morris, C., & Tchistiakova, L. (2016). High-throughput antibody engineering by integrating bioinformatics and mammalian cell display. Journal of Immunology Research, 2016, Article 4216074. https://doi.org/10.1155/2016/4216074
Dondelinger, M., Filée, P., Sauvage, E., Quinting, B., Muyldermans, S., Galleni, M., & Vandevenne, M. (2018). Understanding the significance and implications of antibody numbering and antigen-binding surface/residue definition. Frontiers in Immunology, 9, 2278. https://doi.org/10.3389/fimmu.2018.02278
European Biotechnology. (n.d.). Evolution of antibody humanization and affinity maturation. Retrieved February 3, 2026, from https://european-biotechnology.com/sponsored-publications/evolution-of-antibody-humanization-and-affinity-maturation/
Gordon, G. L., Raybould, M. I. J., Wong, A., & Deane, C. M. (2024). Prospects for the computational humanization of antibodies and nanobodies. Frontiers in Immunology, 15, 1399438. https://doi.org/10.3389/fimmu.2024.1399438
Li, J., Zhang, C., Xia, W., Kan, H. W., Huang, K., Zhang, Y., Peng, Z., Zhu, Q., Ma, H., Jiang, T., & Liu, B. (2025). Significantly enhancing human antibody affinity via deep learning and computational biology-guided single-point mutations. Briefings in Bioinformatics, 26(5), bbaf445. https://doi.org/10.1093/bib/bbaf445
Lisowska, M., Worrall, E. G., Zavadil-Kokas, F., Charlton, K., Murray, E., Mohtar, M. A., Krejcir, R., Hrabal, V., Brydon, J., Gonzalez Urionabarrenetxea, A., Saliba, D. G., Grima, M., Kalathiya, U., Muller, P., Krejci, A., Vojtesek, B., Ball, K., Fahraeus, R., Argyle, D. J., … Hupp, T. R. (2025). The development of a canine single-chain phage antibody library to isolate recombinant antibodies for use in translational cancer research. Cell Reports Methods, 5(3), 101008. https://doi.org/10.1016/j.crmeth.2025.101008
Lu, Y., Huang, W., Li, Y., Xu, Y., Wei, Q., Sha, C., & Guo, P. (2025). Leveraging artificial intelligence in antibody-drug conjugate development: From target identification to clinical translation in oncology. npj Precision Oncology, 9, 374. https://doi.org/10.1038/s41698-025-01159-2
ProteoGenix. (n.d.). Phage display services: Cat. Retrieved February 3, 2026, from https://www.proteogenix.science/custom-antibody/phage-display-services/cat/
ProteoGenix. (n.d.). Dog library for phage display. Retrieved February 3, 2026, from https://www.proteogenix.science/custom-antibody/phage-display-services/dog-library-for-phage-display/
Song, M.-Y., Cho, J., Park, H., Song, Y., Kim, K., Ahn, J.-H., Lee, C.-M., Kim, D. H., & Ko, H.-J. (2025). Discovery and functional characterization of canine PD-L1-targeted antibodies for evaluating antitumor efficacy in a canine osteosarcoma xenograft model. Scientific Reports, 15, 7574. https://doi.org/10.1038/s41598-025-90770-1
Voigt, A., Semenova, T., Yamamoto, J., Etienne, V., & Nguyen, C. Q. (2018). Therapeutic antibody discovery in infectious diseases using single-cell analysis. In Advances in Experimental Medicine and Biology (Vol. 1068, pp. 89–102). Springer. https://doi.org/10.1007/978-981-13-0502-3_8
Wang, J., Zhou, X., Elazab, S. T., Huang, J., & Hsu, W. H. (2025). Current review of monoclonal antibody therapeutics in small animal medicine. Animals, 15(4), 472. https://doi.org/10.3390/ani15040472
Zhang, Y. (2023). Evolution of phage display libraries for therapeutic antibody discovery. mAbs, 15(1), 2213793. https://doi.org/10.1080/19420862.2023.2213793
Zheng, J., Wang, Y., Liang, Q., Cui, L., & Wang, S. (2024). The application of machine learning on antibody discovery and optimization. Molecules, 29(24), 5923. https://doi.org/10.3390/molecules29245923