 Antibody production
Antibody production
Essential antibody library generation and display technologies for the discovery of new reagents and biopharmaceuticals
Library generation is a vital first step in any antibody phage display project for therapeutic applications and others. Libraries exist in different types and formats, and they can be built from different host species. But the success of any antibody discovery project relies, first and foremost, on the underlying quality of the library used for screening, the choice and design of an adequate antigen, and the criteria used for antibody validation.
Evolution of antibody library generation and antibody display methods
The onset of phage display and antibody library generation methods made in vitro antibody discovery possible. These developments also allowed in vitro methods to rival with the decades-old hybridoma technology. Unlike hybridoma, phage display of vast antibody repertoires can be performed with toxic or non-immunogenic antigens making it highly valuable for antivenom drug discovery or for tailoring antibodies to target specific epitopes minimizing cross-reactivity.
Due to the high throughput nature of the technology, antibodies generated by phage display can also be quickly screened for affinity and selectivity (cross-reactivity), two particularly important properties of antibodies used in the clinic as a treatment or diagnostic tools.
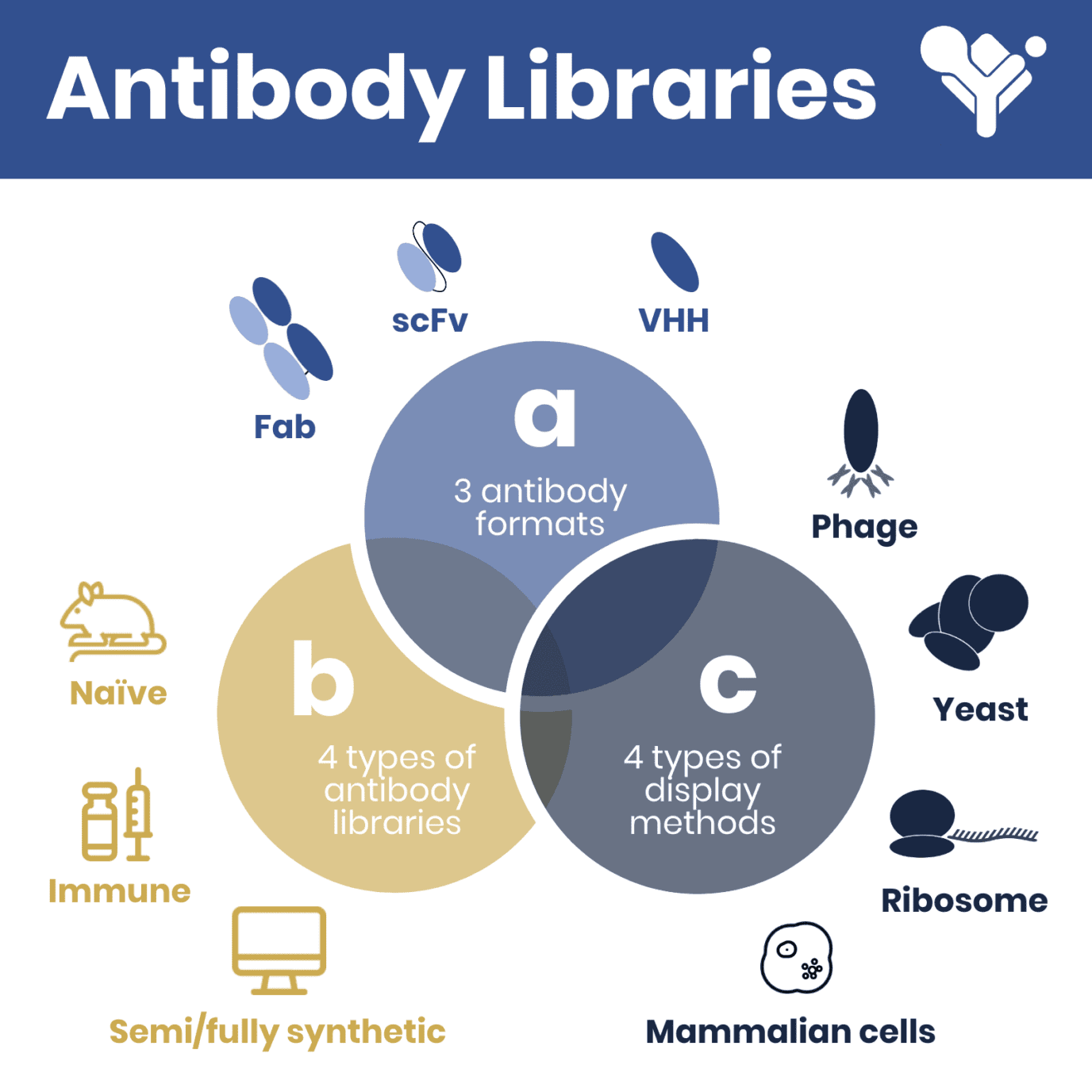
In the past decade, we have witnessed an unparalleled development of antibody library generation and display methods, namely phage, ribosomal, yeast, or mammalian cell surface display. All these methods still share a common limitation: the absence of natural somatic hypermutation (SHM), a biological process responsible for driving affinity maturation of naïve antibodies in the host’s organism. However, several strategies can be used during antibody generation to overcome this limitation.
Overcoming typical challenges in antibody library generation
To overcome the lack of SHM under in vitro conditions, most experts advise focusing on developing the best possible antibody library. Previously, improving the quality solely meant increasing the absolute antibody library diversity. The higher the clonal diversity, the higher the chances of finding the best binders.
However, with the development of in silico methods, libraries are increasingly built by careful analysis of antibody sequences before the experimental screening (e.g. phage, ribosome, yeast, mammalian cell display). By performing a critical assessment of these sequences, researchers can now exclude binders with undesirable physicochemical properties and reduced developability (lower stability, a higher tendency to aggregate, etc.) which allows for a smooth and cost-effective transition to large-scale production. Additionally, “curating” the sequences leads to a reduction of the effective library size and improvement of library’s functionality (absolute library size > functional library size) which, in turn, increases the chances of finding the best binders with a lower number of antibody screening cycles.
Recent advances in display technology have also created other ways to solve the main challenge of in vitro antibody discovery. Antibody’s affinity is typically fine-tuned in vivo by SHM. This process is enzyme-dependent but, so far, difficult to replicate under in vitro conditions.
Conventionally, the weak affinity of naïve binders can be improved by building mutant libraries and performing random or directed mutagenesis on complementarity-determining regions (CDRs). Afterward, mutants with improved affinity can be selected by phage display or other display technologies.
Another solution would be to couple the enzymatic process of SHM with mammalian cell display and fluorescence-activated cell sorting (FACS). This process has been attempted before by using activation-induced cytidine deaminase (AID) in vitro in non-B antibody-producing cells. Studies show that AID-induced mutagenesis in antibody-producing cells can potentially minimize the risks of introduction of unusual or hydrophobic residues (which could hamper antibody developability) and ensure proper protein folding and stability in comparison to in vitro mutagenesis approaches.
Other authors have also described the successful use of CRISPR-Cas9 technologies on low-affinity monoclonal antibodies to drive in vitro affinity maturation. Unlike conventional AID-induced approaches, CRISPR-Cas9 allows researchers to target AID to the immunoglobulin genes. CRISPR-Cas9 guided AID coupled with FACS has already been shown to trigger affinity maturation in recombinant mammalian cells (e.g. HEK) and to produce high-affinity antibodies in only a few rounds of selection.
Principles of antibody library generation
Antibody libraries can be classified according to the format (i.e. scFv, Fab, VHH, etc.) and the source of the antibody repertoire. The Fab format consists of ~50 kDa fragments containing an antigen-binding heavy (VH – CH1) and light chain (VL – CL), while the scFv format consists of ~25 kDa fragments connected by a flexible linker. Fab is typically more stable than scFv, moreover, it’s binding activity is superior, and it can more easily be converted to IgG format.
The VHH format derives solely from heavy-chain antibodies found in camelids (alpaca, camel, llama) and sharks and it is the most stable and lighter (~14 kDa) format typically used for antibody library generation.
Independently from the format, antibody libraries are usually obtained by reverse transcribing and amplifying the antibody binding domain from peripheral blood mononuclear cells or B cells. Subsequently, the heavy and light chains are combined randomly to form either combined scFv or Fab fragments suitable for expression in the chosen display system. This last step is omitted in VHH libraries due to the absence of a light chain.
The source of the cells used for antibody library generation can be naïve hosts (never challenged by a specific antigen) or from immune hosts (challenged by a specific antigen). In these cases, libraries can be classified as naïve or immune, respectively. Due to the technical constraints tied to the low affinity of naïve antibodies, the corresponding naïve libraries should maximize donor number and diversity to ensure the presence of promising binders.
Alternatively, due to the onset of in silico antibody design, antibody library generation has been gradually shifting towards computational methods. Semi-synthetic (a mixture of natural and in silico designed) and fully synthetic antibody libraries are being increasingly used for successful antibody discovery. However, the design of these libraries can be very labor-intensive and complex which has, so far, hampered its widespread use among antibody developers.
Summary
Concluding remarks
Presently, phage display rivals with the hybridoma technology for the generation of antibodies for a multitude of applications. Phage display of vast antibody libraries allows the use of toxic or non-immunogenic antigens and, therefore, it is extremely suitable for antivenom and self-antigen drug discovery projects. Moreover, when the antibody library is of high-quality, phage display can significantly reduce the turnaround time for antibody discovery from 5 months to 6-8 weeks.
However, antibody library generation and display methods still suffer from important limitations, such as the inability to preserve proper pairing information, the high risk of generation immunogenic antibodies, and the difficulty in replicating natural SHM processes to maximize antibody’s affinity.
These drawbacks have driven the development of better antibody library generation and display methods. In recent years, there has been an increased interest in the development of computational methods to increase antibody libraries’ functionality and to improve in vitro affinity maturation. These technologies are further expected to drive the evolution of in vitro discovery methods in the years to come.
- Bailly, M. et al. Predicting Antibody Developability Profiles Through Early Stage Discovery Screening. MAbs. 2020; 12(1): 1743053. doi: 10.1080/19420862.2020.1743053
- Bowers, P. M. et al. Mammalian Cell Display for the Discovery and Optimization of Antibody Therapeutics. Methods. 2014; 65(1):44-56. doi: 10.1016/j.ymeth.2013.06.010
- Devilder, M. C. et al. Ex vivo evolution of human antibodies by CRISPR-X: from a naive B cell repertoire to affinity matured antibodies. BMC Biotechnology. 2019; 19:14. doi: 10.1186/s12896-019-0504-z
- Ponsel, D. et al. High Affinity, Developability and Functional Size: The Holy Grail of Combinatorial Antibody Library Generation. Molecules. 2011; 16(5):3675-700. doi: 10.3390/molecules16053675
- Shim, H. Synthetic approach to the generation of antibody diversity. BMB Rep. 2015; 48(9): 489–494. doi: 10.5483/BMBRep.2015.48.9.120